| Compact overview of VAR_048095 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UniProt ID | Gene | Mutation | Mutation type | Disease | OMIM | dbSNP | |||||||
| 1433S_HUMAN | SFN | M155I | Polymorphism | - | No OMIM entry | rs11542705 | |||||||
| Short stretch summary of VAR_048095 | ||||
|---|---|---|---|---|
| Predictor | Predicted regions overview | Comparison to WT | Stretches in variant | Stretches in WT |
| Tango |  | 0 No change | 2 | 2 |
| Waltz |  | 37 No change | 2 | 1 |
| Limbo |  | 0 No change | 3 | 3 |
| Domain composition of 1433S_HUMAN | ||||
| Database | Domain composition | Residue details | ||
| PFAM |  | 14-3-3 (Residues 3-238) | ||
| SMART |  | 14_3_3 (Residues 3-244) | ||
| Structure stability summary of VAR_048095 | ||||
|---|---|---|---|---|
| Variant | Stability change | Effect | Structure | Stability frequency histogram |
| M155I | 0.52 kcal/mol | Slightly reduced stability |  | 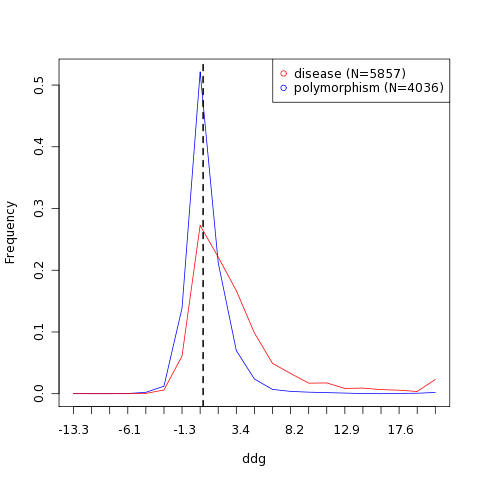 |
TANGO aggregation
| Graphical comparison of TANGO regions in variant and wild type | |||
|---|---|---|---|
| Protein | Predicted regions overview | TANGO regions | Total score |
| Variant |  | 2 | 332 |
| Wild type |  | 2 | 332 |
| TANGO regions in wild type and variant | |||||
|---|---|---|---|---|---|
| Wild type TANGO regions | |||||
| N-term gatekeepers | TANGO region | C-term gatekeepers | Start | End | Score |
| PIRLG | LALNFSVF | HYEIA | 172 | 179 | 20 |
| SYKDS | TLIMQLL | RDNLT | 217 | 223 | 16 |
| Variant protein TANGO regions | |||||
| N-term gatekeepers | TANGO region | C-term gatekeepers | Start | End | Score |
| PIRLG | LALNFSVF | HYEIA | 172 | 179 | 20 |
| SYKDS | TLIMQLL | RDNLT | 217 | 223 | 16 |
| Difference in TANGO aggregation between wild type and variant | |
|---|---|
 | This graph plots the per-residue TANGO aggregation score difference between the wild type protein and this variant. |
| TANGO aggregation profile score plot | |
|---|---|
 | This graph plots the per-residue TANGO aggregation score of the variant protein. From left to right, all residue scores from the N-terminus to the C-terminus are plotted |
WALTZ amylogenicity
| Graphical comparison of WALTZ regions in variant and wild type | |||
|---|---|---|---|
| Protein | Predicted regions overview | WALTZ regions | Total score |
| Variant |  | 2 | 191 |
| Wild type |  | 1 | 154 |
| WALTZ regions in wild type and variant | |||||
|---|---|---|---|---|---|
| Wild type WALTZ regions | |||||
| N-term gatekeepers | WALTZ region | C-term gatekeepers | Start | End | Score |
| PIRLG | LALNFS | VFHYE | 172 | 177 | 19 |
| Variant protein WALTZ regions | |||||
| N-term gatekeepers | WALTZ region | C-term gatekeepers | Start | End | Score |
| SARSA | YQEAID | ISKKE | 151 | 156 | 6 |
| PIRLG | LALNFS | VFHYE | 172 | 177 | 19 |
| Difference in WALTZ amylogenicity between wild type and variant | |
|---|---|
 | This graph plots the per-residue WALTZ amylogenic score difference between the wild type protein and this variant. |
| WALTZ amylogenic profile score plot | |
|---|---|
 | This graph plots the per-residue WALTZ amylogenic score of the variant protein. From left to right, all residue scores from the N-terminus to the C-terminus are plotted |
LIMBO chaperone binding
| Graphical comparison of LIMBO regions in variant and wild type | |||
|---|---|---|---|
| Protein | Predicted regions overview | LIMBO regions | Total score |
| Variant |  | 3 | 2400 |
| Wild type |  | 3 | 2400 |
| LIMBO regions in wild type and variant | |||||
|---|---|---|---|---|---|
| Wild type LIMBO regions | |||||
| N-term gatekeepers | WALTZ region | C-term gatekeepers | Start | End | Score |
| QRAAW | RVLSSIEQ | KSNEE | 60 | 67 | 100 |
| AESRV | FYLKMKGDYYRYLAEV | ATGDD | 119 | 134 | 49 |
| QLLRD | NLTLWTAD | NAGEE | 226 | 233 | 100 |
| Variant protein LIMBO regions | |||||
| N-term gatekeepers | WALTZ region | C-term gatekeepers | Start | End | Score |
| QRAAW | RVLSSIEQ | KSNEE | 60 | 67 | 100 |
| AESRV | FYLKMKGDYYRYLAEV | ATGDD | 119 | 134 | 49 |
| QLLRD | NLTLWTAD | NAGEE | 226 | 233 | 100 |
| Difference in LIMBO chaperone binding between wild type and variant | |
|---|---|
 | This graph plots the per-residue LIMBO chaperone binding difference between the wild type protein and this variant. |
| LIMBO chaperone binding score plot | |
|---|---|
 | This graph plots the per-residue LIMBO chaperone binding score of the variant protein. From left to right, all residue scores from the N-terminus to the C-terminus are plotted |
FOLDX structural profile
Functional sites, structural features and cellular processing
| Functional sites and structural features | |
|---|---|
| Feature | This residue |
| Catalytic site | No |
| Secondary structure | HELIX |
| Transmembrane topology | No TM regions in this protein |
| Cellular processing | |
|---|---|
| Feature | Affected by variant |
| Signal peptide | No |
| Farnesylation anchor | No |
| N-Myristoylation anchor | No |
| Geranylgeranyl transferase Type 1 anchor | No |
| Geranylgeranyl transferase Type 2 anchor | No |
| Glycosylphosphatidylinositol (GPI) anchor | No |
| Peroxisomal targeting signal PTS1 | No |
| Subcellular location | No |
All mutations from 1433S_HUMAN
1 mutations listed
| Variant | UniProt ID | Mutation | Disease | Mutation Type | dTANGO | dWALTZ | dLIMBO | ddG |
|---|---|---|---|---|---|---|---|---|
| VAR_048095 | 1433S_HUMAN | M155I | - | Polymorphism | 0 | 37 | 0 | 0.52 |








