| Compact overview of VAR_004335 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UniProt ID | Gene | Mutation | Mutation type | Disease | OMIM | dbSNP | |||||||
| 1A02_HUMAN | HLA-A | D54N | Polymorphism | - | No OMIM entry | No dbSNP entry | |||||||
| Short stretch summary of VAR_004335 | ||||
|---|---|---|---|---|
| Predictor | Predicted regions overview | Comparison to WT | Stretches in variant | Stretches in WT |
| Tango |  | 81 Increased | 6 | 6 |
| Waltz |  | 0 No change | 2 | 2 |
| Limbo |  | 0 No change | 5 | 5 |
| Domain composition of 1A02_HUMAN | ||||
| Database | Domain composition | Residue details | ||
| PFAM |  | MHC_I (Residues 25-203), C1-set (Residues 212-295), MHC_I_C (Residues 336-364) | ||
| SMART |  | IGc1 (Residues 222-293) | ||
| Structure stability summary of VAR_004335 | ||||
|---|---|---|---|---|
| Variant | Stability change | Effect | Structure | Stability frequency histogram |
| D54N | -0.32 kcal/mol | No effect on stability |  |  |
TANGO aggregation
| Graphical comparison of TANGO regions in variant and wild type | |||
|---|---|---|---|
| Protein | Predicted regions overview | TANGO regions | Total score |
| Variant |  | 6 | 3036 |
| Wild type |  | 6 | 2956 |
| TANGO regions in wild type and variant | |||||
|---|---|---|---|---|---|
| Wild type TANGO regions | |||||
| N-term gatekeepers | TANGO region | C-term gatekeepers | Start | End | Score |
| VMAPR | TLVLLLSGALALT | QTWAG | 8 | 20 | 51 |
| HSMRY | FFTSV | SRPGR | 32 | 36 | 6 |
| RGEPR | FIAVGYV | DDTQF | 46 | 52 | 77 |
| FYPAE | ITLTW | QRDGE | 237 | 241 | 13 |
| GTFQK | WAAVV | VPSGQ | 268 | 272 | 11 |
| QPTIP | IVGIIAGLVLFGAVITGAVVAAVMW | RRKSS | 308 | 332 | 63 |
| Variant protein TANGO regions | |||||
| N-term gatekeepers | TANGO region | C-term gatekeepers | Start | End | Score |
| VMAPR | TLVLLLSGALALT | QTWAG | 8 | 20 | 51 |
| HSMRY | FFTSV | SRPGR | 32 | 36 | 6 |
| RGEPR | FIAVGYV | DNTQF | 46 | 52 | 89 |
| FYPAE | ITLTW | QRDGE | 237 | 241 | 13 |
| GTFQK | WAAVV | VPSGQ | 268 | 272 | 11 |
| QPTIP | IVGIIAGLVLFGAVITGAVVAAVMW | RRKSS | 308 | 332 | 63 |
| Difference in TANGO aggregation between wild type and variant | |
|---|---|
 | This graph plots the per-residue TANGO aggregation score difference between the wild type protein and this variant. |
| TANGO aggregation profile score plot | |
|---|---|
 | This graph plots the per-residue TANGO aggregation score of the variant protein. From left to right, all residue scores from the N-terminus to the C-terminus are plotted |
WALTZ amylogenicity
| Graphical comparison of WALTZ regions in variant and wild type | |||
|---|---|---|---|
| Protein | Predicted regions overview | WALTZ regions | Total score |
| Variant |  | 2 | 186 |
| Wild type |  | 2 | 187 |
| WALTZ regions in wild type and variant | |||||
|---|---|---|---|---|---|
| Wild type WALTZ regions | |||||
| N-term gatekeepers | WALTZ region | C-term gatekeepers | Start | End | Score |
| FLRGY | HQYAYD | GKDYI | 138 | 143 | 22 |
| TLRCW | ALSFYP | AEITL | 229 | 234 | 6 |
| Variant protein WALTZ regions | |||||
| N-term gatekeepers | WALTZ region | C-term gatekeepers | Start | End | Score |
| FLRGY | HQYAYD | GKDYI | 138 | 143 | 22 |
| TLRCW | ALSFYP | AEITL | 229 | 234 | 6 |
| Difference in WALTZ amylogenicity between wild type and variant | |
|---|---|
 | This graph plots the per-residue WALTZ amylogenic score difference between the wild type protein and this variant. |
| WALTZ amylogenic profile score plot | |
|---|---|
 | This graph plots the per-residue WALTZ amylogenic score of the variant protein. From left to right, all residue scores from the N-terminus to the C-terminus are plotted |
LIMBO chaperone binding
| Graphical comparison of LIMBO regions in variant and wild type | |||
|---|---|---|---|
| Protein | Predicted regions overview | LIMBO regions | Total score |
| Variant |  | 5 | 3189 |
| Wild type |  | 5 | 3189 |
| LIMBO regions in wild type and variant | |||||
|---|---|---|---|---|---|
| Wild type LIMBO regions | |||||
| N-term gatekeepers | WALTZ region | C-term gatekeepers | Start | End | Score |
| MAPRT | LVLLLSGA | LALTQ | 9 | 16 | 100 |
| VGSDW | RFLRGYHQ | YAYDG | 132 | 139 | 66 |
| TCVEW | LRRYLENG | KETLQ | 192 | 199 | 87 |
| FYPAE | ITLTWQRD | GEDQT | 237 | 244 | 46 |
| GLPKP | LTLRWEPS | SQPTI | 294 | 301 | 98 |
| Variant protein LIMBO regions | |||||
| N-term gatekeepers | WALTZ region | C-term gatekeepers | Start | End | Score |
| MAPRT | LVLLLSGA | LALTQ | 9 | 16 | 100 |
| VGSDW | RFLRGYHQ | YAYDG | 132 | 139 | 66 |
| TCVEW | LRRYLENG | KETLQ | 192 | 199 | 87 |
| FYPAE | ITLTWQRD | GEDQT | 237 | 244 | 46 |
| GLPKP | LTLRWEPS | SQPTI | 294 | 301 | 98 |
| Difference in LIMBO chaperone binding between wild type and variant | |
|---|---|
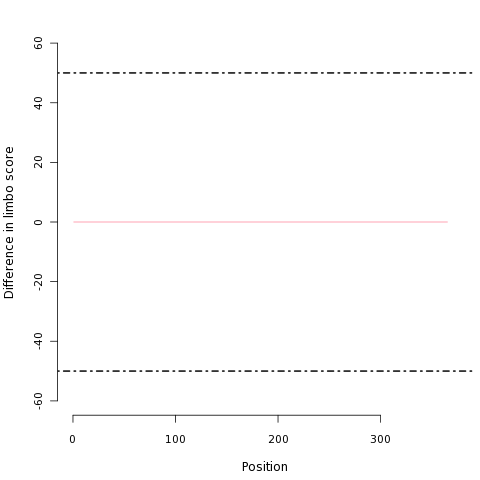 | This graph plots the per-residue LIMBO chaperone binding difference between the wild type protein and this variant. |
| LIMBO chaperone binding score plot | |
|---|---|
 | This graph plots the per-residue LIMBO chaperone binding score of the variant protein. From left to right, all residue scores from the N-terminus to the C-terminus are plotted |
FOLDX structural profile
Functional sites, structural features and cellular processing
| Functional sites and structural features | |
|---|---|
| Feature | This residue |
| Catalytic site | No |
| Secondary structure | No secondary structure information available |
| Transmembrane topology | outside |
| Cellular processing | |
|---|---|
| Feature | Affected by variant |
| Signal peptide | No |
| Farnesylation anchor | No |
| N-Myristoylation anchor | No |
| Geranylgeranyl transferase Type 1 anchor | No |
| Geranylgeranyl transferase Type 2 anchor | No |
| Glycosylphosphatidylinositol (GPI) anchor | No |
| Peroxisomal targeting signal PTS1 | No |
| Subcellular location | No |
All mutations from 1A02_HUMAN
22 mutations listed
| Variant | UniProt ID | Mutation | Disease | Mutation Type | dTANGO | dWALTZ | dLIMBO | ddG |
|---|---|---|---|---|---|---|---|---|
| VAR_016730 | 1A02_HUMAN | W191G | - | Polymorphism | 0 | -1 | 8 | 1.99 |
| VAR_004350 | 1A02_HUMAN | A260E | - | Polymorphism | 0 | 0 | 0 | 1.31 |
| VAR_004349 | 1A02_HUMAN | T187E | - | Polymorphism | 0 | -1 | 0 | -1.03 |
| VAR_016728 | 1A02_HUMAN | H98D | - | Polymorphism | 0 | 0 | 0 | 1.96 |
| VAR_016729 | 1A02_HUMAN | E190D | - | Polymorphism | 0 | -1 | 0 | 0.41 |
| VAR_004348 | 1A02_HUMAN | L180Q | - | Polymorphism | 0 | 1 | 0 | 1.75 |
| VAR_016727 | 1A02_HUMAN | H94Q | - | Polymorphism | 0 | 0 | 0 | -0.31 |
| VAR_016726 | 1A02_HUMAN | A65G | - | Polymorphism | 0 | 0 | 0 | 0.23 |
| VAR_004344 | 1A02_HUMAN | M162K | - | Polymorphism | 1 | 0 | 0 | -0.28 |
| VAR_004347 | 1A02_HUMAN | L180W | - | Polymorphism | 0 | -1 | 0 | -0.56 |
| VAR_004345 | 1A02_HUMAN | A173T | - | Polymorphism | 0 | 0 | 0 | 0.3 |
| VAR_004346 | 1A02_HUMAN | V176E | - | Polymorphism | 0 | 0 | 0 | 0.69 |
| VAR_004337 | 1A02_HUMAN | K90N | - | Polymorphism | -1 | 0 | 0 | -0.06 |
| VAR_004343 | 1A02_HUMAN | W131G | - | Polymorphism | 0 | -1 | -529 | -0.56 |
| VAR_004338 | 1A02_HUMAN | T97I | - | Polymorphism | 0 | 0 | 0 | -0.54 |
| VAR_004339 | 1A02_HUMAN | V119L | - | Polymorphism | 0 | 0 | 0 | -0.63 |
| VAR_004340 | 1A02_HUMAN | R121M | - | Polymorphism | 0 | 0 | 1 | 0.57 |
| VAR_004341 | 1A02_HUMAN | Y123C | - | Polymorphism | 0 | 0 | 0 | 2.15 |
| VAR_004342 | 1A02_HUMAN | Y123F | - | Polymorphism | 0 | 0 | 0 | 0.22 |
| VAR_004335 | 1A02_HUMAN | D54N | - | Polymorphism | 81 | 0 | 0 | -0.32 |
| VAR_004336 | 1A02_HUMAN | Q67R | - | Polymorphism | 1 | 0 | 786 | 0.48 |
| VAR_004334 | 1A02_HUMAN | F33Y | - | Polymorphism | -24 | -3 | 0 | 0.92 |








