| Compact overview of VAR_004374 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UniProt ID | Gene | Mutation | Mutation type | Disease | OMIM | dbSNP | |||||||
| 1A32_HUMAN | HLA-A | L180Q | Polymorphism | - | No OMIM entry | No dbSNP entry | |||||||
| Short stretch summary of VAR_004374 | ||||
|---|---|---|---|---|
| Predictor | Predicted regions overview | Comparison to WT | Stretches in variant | Stretches in WT |
| Tango |  | 0 No change | 5 | 5 |
| Waltz |  | 2 No change | 2 | 2 |
| Limbo |  | 0 No change | 6 | 6 |
| Domain composition of 1A32_HUMAN | ||||
| Database | Domain composition | Residue details | ||
| PFAM |  | MHC_I (Residues 25-203), C1-set (Residues 212-295), MHC_I_C (Residues 336-364) | ||
| SMART |  | IGc1 (Residues 222-293) | ||
| Structure stability summary of VAR_004374 | ||||
|---|---|---|---|---|
| Variant | Stability change | Effect | Structure | Stability frequency histogram |
| L180Q | 1.13 kcal/mol | Reduced stability |  | 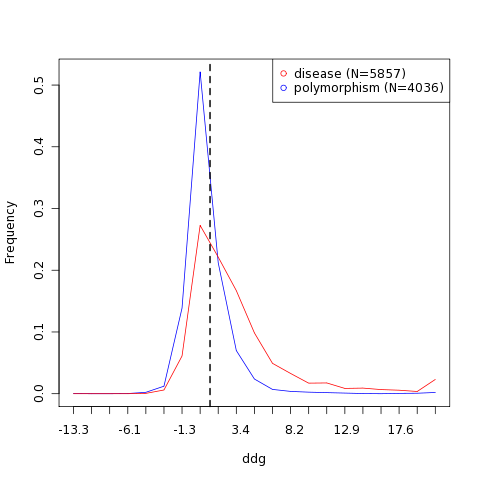 |
TANGO aggregation
| Graphical comparison of TANGO regions in variant and wild type | |||
|---|---|---|---|
| Protein | Predicted regions overview | TANGO regions | Total score |
| Variant |  | 5 | 3116 |
| Wild type |  | 5 | 3116 |
| TANGO regions in wild type and variant | |||||
|---|---|---|---|---|---|
| Wild type TANGO regions | |||||
| N-term gatekeepers | TANGO region | C-term gatekeepers | Start | End | Score |
| VMAPR | TLLLLLLGALALTQ | TWAGS | 8 | 21 | 70 |
| HSMRY | FFTSV | SRPGR | 32 | 36 | 6 |
| RGEPR | FIAVGYV | DDTQF | 46 | 52 | 77 |
| FYPAE | ITLTW | QRDGE | 237 | 241 | 13 |
| QPTIP | IVGIIAGLVLFGAMFAGAVVAAV | RWRRK | 308 | 330 | 64 |
| Variant protein TANGO regions | |||||
| N-term gatekeepers | TANGO region | C-term gatekeepers | Start | End | Score |
| VMAPR | TLLLLLLGALALTQ | TWAGS | 8 | 21 | 70 |
| HSMRY | FFTSV | SRPGR | 32 | 36 | 6 |
| RGEPR | FIAVGYV | DDTQF | 46 | 52 | 77 |
| FYPAE | ITLTW | QRDGE | 237 | 241 | 13 |
| QPTIP | IVGIIAGLVLFGAMFAGAVVAAV | RWRRK | 308 | 330 | 64 |
| Difference in TANGO aggregation between wild type and variant | |
|---|---|
 | This graph plots the per-residue TANGO aggregation score difference between the wild type protein and this variant. |
| TANGO aggregation profile score plot | |
|---|---|
 | This graph plots the per-residue TANGO aggregation score of the variant protein. From left to right, all residue scores from the N-terminus to the C-terminus are plotted |
WALTZ amylogenicity
| Graphical comparison of WALTZ regions in variant and wild type | |||
|---|---|---|---|
| Protein | Predicted regions overview | WALTZ regions | Total score |
| Variant |  | 2 | 335 |
| Wild type |  | 2 | 334 |
| WALTZ regions in wild type and variant | |||||
|---|---|---|---|---|---|
| Wild type WALTZ regions | |||||
| N-term gatekeepers | WALTZ region | C-term gatekeepers | Start | End | Score |
| ESLRI | ALRYYN | QSEAG | 105 | 110 | 47 |
| TLRCW | ALSFYP | AEITL | 229 | 234 | 6 |
| Variant protein WALTZ regions | |||||
| N-term gatekeepers | WALTZ region | C-term gatekeepers | Start | End | Score |
| ESLRI | ALRYYN | QSEAG | 105 | 110 | 47 |
| TLRCW | ALSFYP | AEITL | 229 | 234 | 6 |
| Difference in WALTZ amylogenicity between wild type and variant | |
|---|---|
 | This graph plots the per-residue WALTZ amylogenic score difference between the wild type protein and this variant. |
| WALTZ amylogenic profile score plot | |
|---|---|
 | This graph plots the per-residue WALTZ amylogenic score of the variant protein. From left to right, all residue scores from the N-terminus to the C-terminus are plotted |
LIMBO chaperone binding
| Graphical comparison of LIMBO regions in variant and wild type | |||
|---|---|---|---|
| Protein | Predicted regions overview | LIMBO regions | Total score |
| Variant |  | 6 | 2913 |
| Wild type |  | 6 | 2913 |
| LIMBO regions in wild type and variant | |||||
|---|---|---|---|---|---|
| Wild type LIMBO regions | |||||
| N-term gatekeepers | WALTZ region | C-term gatekeepers | Start | End | Score |
| MAPRT | LLLLLLGAL | ALTQT | 9 | 17 | 89 |
| RESLR | IALRYY | NQSEA | 104 | 109 | 7 |
| AGSHT | IQMMYGCD | VGPDG | 119 | 126 | 25 |
| TCVEW | LRRYLENG | KETLQ | 192 | 199 | 87 |
| FYPAE | ITLTWQRD | GEDQT | 237 | 244 | 46 |
| GLPKP | LTLRWEPS | SQPTI | 294 | 301 | 98 |
| Variant protein LIMBO regions | |||||
| N-term gatekeepers | WALTZ region | C-term gatekeepers | Start | End | Score |
| MAPRT | LLLLLLGAL | ALTQT | 9 | 17 | 89 |
| RESLR | IALRYY | NQSEA | 104 | 109 | 7 |
| AGSHT | IQMMYGCD | VGPDG | 119 | 126 | 25 |
| TCVEW | LRRYLENG | KETLQ | 192 | 199 | 87 |
| FYPAE | ITLTWQRD | GEDQT | 237 | 244 | 46 |
| GLPKP | LTLRWEPS | SQPTI | 294 | 301 | 98 |
| Difference in LIMBO chaperone binding between wild type and variant | |
|---|---|
 | This graph plots the per-residue LIMBO chaperone binding difference between the wild type protein and this variant. |
| LIMBO chaperone binding score plot | |
|---|---|
 | This graph plots the per-residue LIMBO chaperone binding score of the variant protein. From left to right, all residue scores from the N-terminus to the C-terminus are plotted |
FOLDX structural profile
Functional sites, structural features and cellular processing
| Functional sites and structural features | |
|---|---|
| Feature | This residue |
| Catalytic site | No |
| Secondary structure | No secondary structure information available |
| Transmembrane topology | outside |
| Cellular processing | |
|---|---|
| Feature | Affected by variant |
| Signal peptide | No |
| Farnesylation anchor | No |
| N-Myristoylation anchor | No |
| Geranylgeranyl transferase Type 1 anchor | No |
| Geranylgeranyl transferase Type 2 anchor | No |
| Glycosylphosphatidylinositol (GPI) anchor | No |
| Peroxisomal targeting signal PTS1 | No |
| Subcellular location | No |
All mutations from 1A32_HUMAN
13 mutations listed
| Variant | UniProt ID | Mutation | Disease | Mutation Type | dTANGO | dWALTZ | dLIMBO | ddG |
|---|---|---|---|---|---|---|---|---|
| VAR_016832 | 1A32_HUMAN | P129S | - | Polymorphism | 0 | 0 | 0 | 1.78 |
| VAR_056285 | 1A32_HUMAN | I166T | - | Polymorphism | 0 | -1 | 0 | 1.98 |
| VAR_056284 | 1A32_HUMAN | N151K | - | Polymorphism | 0 | 0 | 2 | -1.11 |
| VAR_056283 | 1A32_HUMAN | R89G | - | Polymorphism | -1 | 0 | 0 | 1.36 |
| VAR_016835 | 1A32_HUMAN | Q168K | - | Polymorphism | 1 | -1 | 0 | -0.75 |
| VAR_016833 | 1A32_HUMAN | L133F | - | Polymorphism | 0 | 0 | 0 | -1.27 |
| VAR_016834 | 1A32_HUMAN | Q138R | - | Polymorphism | 1 | 0 | 9 | -0.54 |
| VAR_004374 | 1A32_HUMAN | L180Q | - | Polymorphism | 0 | 2 | 0 | 1.13 |
| VAR_016831 | 1A32_HUMAN | M121I | - | Polymorphism | 11 | 1 | -97 | 0.03 |
| VAR_016657 | 1A32_HUMAN | S101N | - | Polymorphism | 0 | 1 | 10 | -0.59 |
| VAR_016837 | 1A32_HUMAN | E185D | - | Polymorphism | 0 | 0 | 0 | 1.47 |
| VAR_016836 | 1A32_HUMAN | V176E | - | Polymorphism | 0 | 0 | 0 | 0.89 |
| VAR_004373 | 1A32_HUMAN | R175H | - | Polymorphism | -1 | 0 | 0 | 0.28 |








